Becker's World of the Cell (9th Edition)
9th Edition
ISBN: 9780321934925
Author: Jeff Hardin, Gregory Paul Bertoni
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 17, Problem 17.1PS
Meselson and Stahl Revisited. For each of the alternative models of
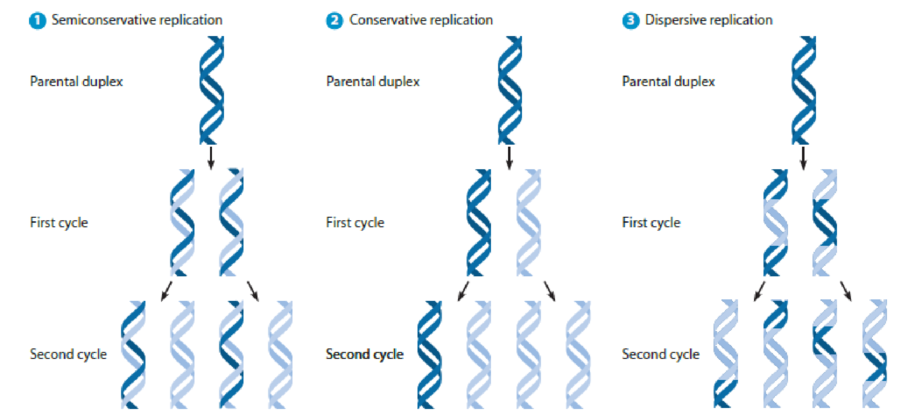
Figure 17-3 Alternative Models for DNA Replication. The results expected for two cycles of DNA replication are shown for each model, which were tested in the experiments of Meselson and Stahl.
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Recall the semi discontinuity in DNA replication. Is it biologically possible for DNA to undergo replication in vivo, without the lagging and the leading strands? In PCR, we also start with an antiparallel strand of DNA. Does this also mean that replication is semidiscontinuous in in vitro replication? PLEASE EXPLAIN COMPLETELY AND NOT JUST COPY PASTE ANSWERS FROM GOOGLE.
Calculating Transformation Efficiency
Transformation Efficiency is defined as the number of colony forming units (cfu) which
would be produced by transforming 1 ug of plasmid DNA into a given volume of competent
cells.
Transformation Efficiency (TE) = number of cfu/ug DNA
You have just transformed 1 ul (100 pg/ul) of control pWasabi (plasmid) DNA into 50 µl of
E. coli DH5a by the heat-shock method. You then add 950 µl of SOC medium to outgrow the
bacteria. Of this, you plated 50 µl onto a LB plate containing ampicillin. After overnight
incubation, you observed that there are 150 colony forming units (cfu) on the plate. Calculate
the transformation efficiency (show your step-wise calculations below).
Which statements are true? Explain why or why not.1 The different cells in your body rarely havegenomes with the identical nucleotide sequence.2 In E. coli, where the replication fork travels at 500nucleotide pairs per second, the DNA ahead of the fork—in the absence of topoisomerase—would have to rotate atnearly 3000 revolutions per minute.3 In a replication bubble, the same parental DNAstrand serves as the template strand for leading-strandsynthesis in one replication fork and as the template forlagging-strand synthesis in the other fork.4 When bidirectional replication forks from adja-cent origins meet, a leading strand always runs into a lag-ging strand.5 DNA repair mechanisms all depend on the exis-tence of two copies of the genetic information, one in eachof the two homologous chromosomes
Chapter 17 Solutions
Becker's World of the Cell (9th Edition)
Ch. 17 - The theoretical amplification accomplished by n...Ch. 17 - Bacterial replication and that in typical...Ch. 17 - Nonhomologous end-joining and synthesis-dependent...Ch. 17 - Prob. 17.3CCCh. 17 - Meselson and Stahl Revisited. For each of the...Ch. 17 - DNA Replication. Sketch a replication fork of...Ch. 17 - More DNA Replication. The following are...Ch. 17 - QUANTITATIVE Still More DNA Replication. Suppose...Ch. 17 - The Minimal Chromosome. To enable it to be...Ch. 17 - Prob. 17.6PS
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- Please help me solve this problem. I am really having a hard time understanding this lesson. Please help. Kindly provide all the necessary information to this problem. Thank you! Please answer numbers 1-5 determine what amino acid will be formed from the given DNA strand below: 3’ T A C A T G C C G A A T G C C 5’ Note: Prepare the partner strand of this DNA. Discuss how will replication happen by mentioning the enzyme needed then transcribe to form mRNA. Discuss what will happen to mRNA, then translate, mentioning the anticodon to be used. Look at the genetic code to know what amino acid will become part of the polypeptide chain. 1. Partner DNA strand 2. the mRNA strand 3. The tRNA 4. the formed amino acids 5. the discussion of the entire procedurearrow_forwardHuman Genome Replication Rate Assume DNA replication proceeds at a rate of 100 base pairs per second in human cells and origins of replication occur every 300 kbp. Assume also that human DNA polymerases are highly processive and only two molecules of DNA polymerase arc needed per replication fork. How long would it take to replicate the entire diploid human genome? How many molecules of DNA polymerase does each cell need to carry out this task?arrow_forwardRECALL Single-stranded regions of DNA are attacked by nucleases in the cell, yet portions of DNA are in a single-stranded form during the replication process. Explain.arrow_forward
- RECALL What are the functions of the gyrase, primase, and ligase enzymes in DNA replication?arrow_forwardCalculating Transformation Efficiency Transformation Efficiency is defined as the number of colony forming units (cfu) which would be produced by transforming 1 ug of plasmid DNA into a given volume of competent cells. Transformation Efficiency (TE) = number of cfu/ug DNA You have just transformed 1 ul (100 pg/ul) of control pWasabi (plasmid) DNA into 50 ul of E. coli DH5oa by the heat-shock method. You then add 950 µl of SOC medium to outgrow the bacteria. Of this, you plated 50 µl onto a LB plate containing ampicillin. After overnight incubation, you observed that there are 150 colony forming units (cfu) on the plate. Calculate the transformation efficiency (show your step-wise calculations below).arrow_forwardPstl. EcoRI Origin of replication (ori) Ampicillin Tetracycline resistance resistance (Amp) (Tet") pBR322 (4,361 bp) BamHI Pvull Sall Recombinant Plasmid DNA Bacterial cell contains... No plasmid DNA pBR322 (no insert) Recombinant plasmid 00,000. Host DNA Transformation of E. coli cells +AMP plate pBR322 Figure 7-5 +TET plate Based on the recombinant plasmid growth pattern (bottom row of blue table), which of the depicted plasmid's restriction sites was used to prepare this sample? Explain how you can tell.arrow_forward
- May you please help me with this? A sample of purified DNA was incubated with deoxyribonuclease (DNAse) at 37oC. An aliquot was removed from the reaction mixture every minute for 5 minutes and the A260 recorded. The following data were obtained. Time (min) A260 0 0.60 1 0.64 2 0.67 3 0.70 4 0.72 5 0.73 Describe the action of deoxyribonuclease on DNA and explain the increase in A260.arrow_forward2)(recall) During DNA replication one nucleotide strand is used as a template for polymerization of another. What WOULD BE THE NUCLEOTIDE (?) in the newly synthesized complimentary DNA chain across from the nucleotide T? template ATGCAGTTCCAGTCGGTA ATG new strand (?) A)U B)A C T or G E) none of the above D) T 3)(recall) Replication: what would be the CORRECT ORDER OF NUCLEOTIDES (? ? ? ? ? ?) in the newly synthesized complimentary growing strand corresponding highlighted sequence of "old" strand? "old" DNA strand ATGCACTTGGAGTCGG TAATG - - - ? ? ? ? ? ? - - new DNA strand --- D) GAACCT A)CTTGGA BGAACCA C)GAACCU E) none of abovearrow_forwardDraw a replication bubble. Be sure to label the directionality of all strands of DNA. For one of the two replication forks, draw and label all of the proteins required the text describes as being important for DNA synthesis, and label the leading and lagging strands.arrow_forward
- 4a in context to taking genomic DNA from eukaryotic cells and randomly shearing it into pieces of a constant size, why do some of the genomic DNA fragments re-nature so much more quickly than other fragmentsarrow_forwardnand portable. 6.5 mm micro-edge 16405U processon USE YOUR SMARTPHONE w micro-edge display design See dnclamers on product bo Revi Vide Fea Sameness and Variety (Mitosis and Meiosis) 161 Spe Example Sup SCAN Expressed using apples, DNA replication looks like this. Go look at the model of DNA on the demonstration table. We have been presenting DNA as a straight ladder, but actually it is twisted on itself like a spiral staircase. This shape is called a heliz. Observe The Chromosome Normally DNA exists as loose strands (chromatin) in the nucleus of a cell. This nuclear DNA sends a message (RNA) to the ribosomes where protein and enzymes are synthesized. When stretched out, the length of one DNA molecule in a human cell is almost 4 cm. However, the cell itself is but a tiny fraction of that size. During cell reproduction the DNA must be able to move around. So it shortens its length by tightly coiling up. In doing so, the DNA strands become wider and are visible under a microscope.…arrow_forwardPick a plasmid . What was its approximate transformation? Express it in # colonies per microgram of DNA transformed. Assume the original DNA was about .001 ug/ul . Count how many colonies you got on one plate (or estimate that number) and figure out how much of the total solution you plated on that plate. Multiply by all the plates, if you plated all of it. OR, if you only plated some of it, figure out how many colonies you would have gotten had you plated all of it. Divide by the number of ug used.arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning
Genome Annotation, Sequence Conventions and Reading Frames; Author: Loren Launen;https://www.youtube.com/watch?v=MWvYgGyqVys;License: Standard Youtube License